Covary protocol portfolio is now available.
A translation-aware framework for alignment-free phylogenetics using machine learning
Protocols
Features
Covary is a computational framework designed for large-scale biological sequence analysis, powered by TIPs-VF.
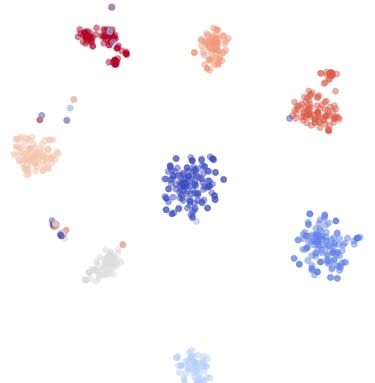

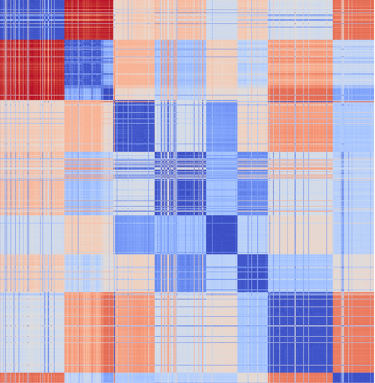
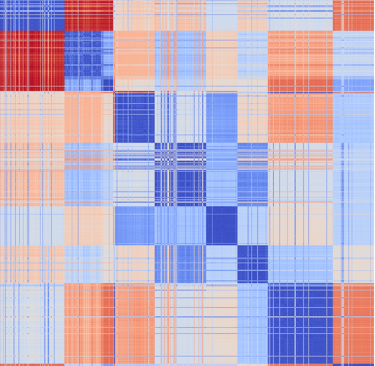
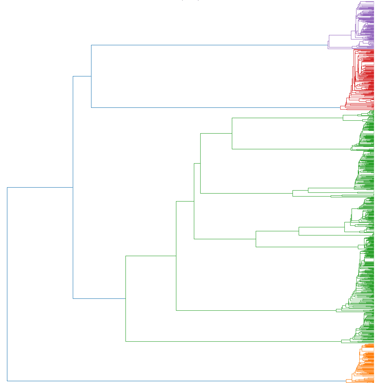
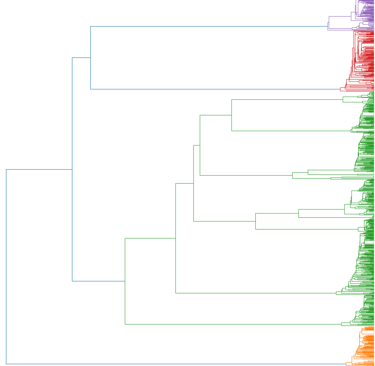
Alignment-free analysis
Translation-aware embeddings
Distance-based clustering
Phylogenetic resolution
Toolkits
Phylogenomic tools, data processing workflows and simulation pipelines that are built for Covary.
Mutagen-PX
A lightweight Python toolkit that simulates tumor-specific gene sequence profiles by applying patient mutation data from TCGA cohorts to a reference sequence. It recreates/simulates “mutated” FASTA outputs per patient, allowing comparative genomics and downstream mutational analyses.
Seed Aligner
A computationally-optimized tool that detects a common seed region across genetic sequences and reorders them to start at the same point. It standardizes FASTA inputs for Covary without performing full multiple sequence alignment.
Covary-encoder
A k-mer-derived, non-overlapping, and frequency-independent encoding logic. It represents genetic sequences based on the relative proximity and directional alignment of k-mer attributes while incorporating sequence, length, positional, and translation awareness.
Usage
Covary leverages alignment-free, translation-aware embeddings to compare, cluster, and analyze genetic sequences, enabling insights into phylogenetic relationships, functional divergence, and taxonomic resolution.



Perform large-scale phylogenomic analyses
Embed sequences with translation-aware context
Run fast, scalable exploratory workflows
Analyze sequences without alignment or gaps
Use Cases
Covary is suitable for use in solving a number of research questions.
Taxonomic studies
Covary provides alignment-free, translation-aware comparisons that help distinguish species, infer relatedness, and support phylogenetic analyses even across highly divergent sequences.

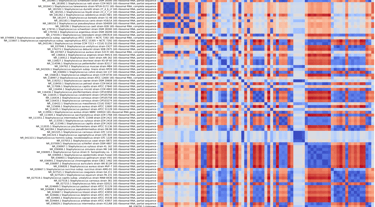
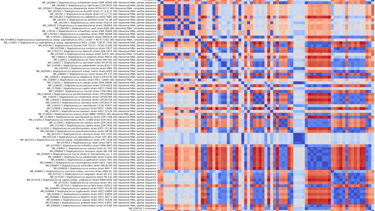
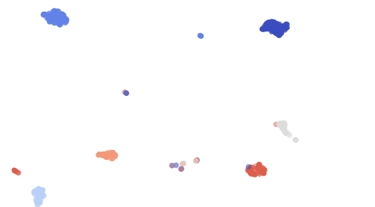

Pathogen detection
Covary can rapidly screen genetic sequences from clinical or environmental samples to identify viral, bacterial, or fungal pathogens without requiring multiple sequence alignment.
Metagenomic exploration
Covary enables fast, large-scale profiling of mixed microbial communities, helping researchers uncover taxonomic composition, detect rare organisms, and analyze functional divergence within complex datasets.
Publications
Covary: A translation-aware framework for alignment-free phylogenetics using machine learning. bioRxiv, 2025-11. https://doi.org/10.1101/2025.11.13.687960
Rapid Phylogenomic Analysis of Thousands Outbreak‐Causing Viral Genomes Using Covary.Preprints, 2025-12. https://doi.org/10.20944/preprints202512.1970.v1
Contact us
Covary is powered by:




© 2025 Marvin De los Santos
All rights reserved

Privacy | Terms | License Notice | Request Access | Procurement
Part of ChordexBio – algorithms empowering lives
